Loading...
Searching...
No Matches
testLJ.cpp
An example demonstrating the computation of Piola and Cauchy stress tensors using hardy and virial spherical averaging domains, and 1D and 2D grids
- Open the configuration file (*.data in MDStressLab format) and read the number of particles
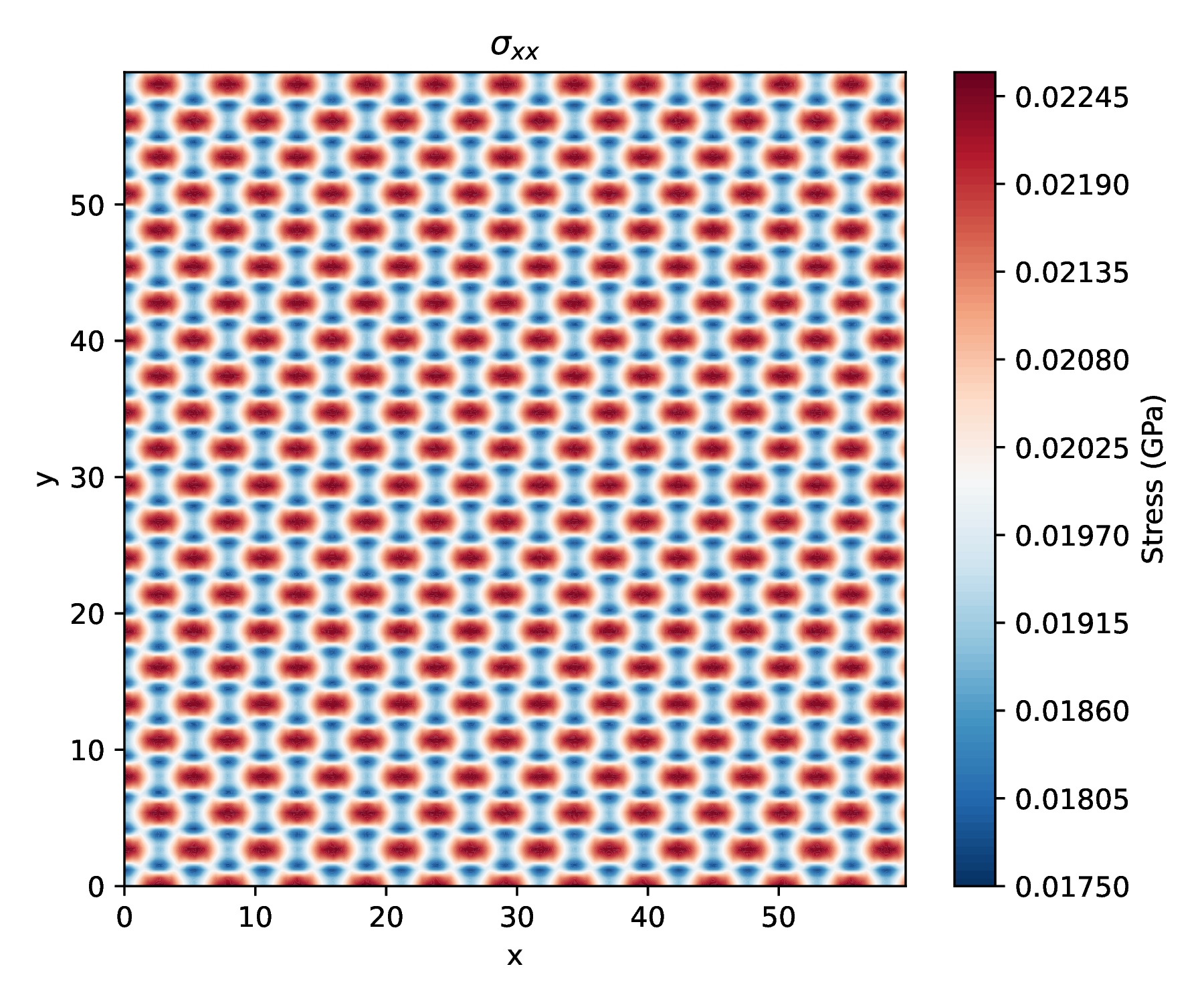
- Initialize the BoxConfiguration and read the reference and current atomic coordinates from the configuration file. The reference configuration is an fcc Ar crystal in the relaxed state. The deformed/current configuration is the reference crystal strained in the \(y\)-direction. Represents a particle configuration including simulation box information.Definition BoxConfiguration.h:33void read(std::string configFileName, int referenceAndFinal)A function to read the properties of atoms from a file in a MDStressLab format.Definition BoxConfiguration.cpp:33
- Initialize the Kim model
- Create 1D and 2D grids
- Construct hardy and virial MethodSphere objects Implements radially symmetric weighting functions (Hardy, Virial) and its associated bond function fo...Definition MethodSphere.h:29
- Construct Stress objects using the MethodSphere and Grid objects Definition Stress.h:34
- Calculate stresses. The following snippet shows that stresses can be calculated all at once or with various combinations. int calculateStress(const BoxConfiguration &body, Kim &kim, std::tuple<> stress, const bool &projectForces=false)Definition calculateStress.cpp:26
- Write stresses
- We compare our results with the exact results for unit testing purposes.
Visual comparison
- See Visualization Utilities for contour plotting
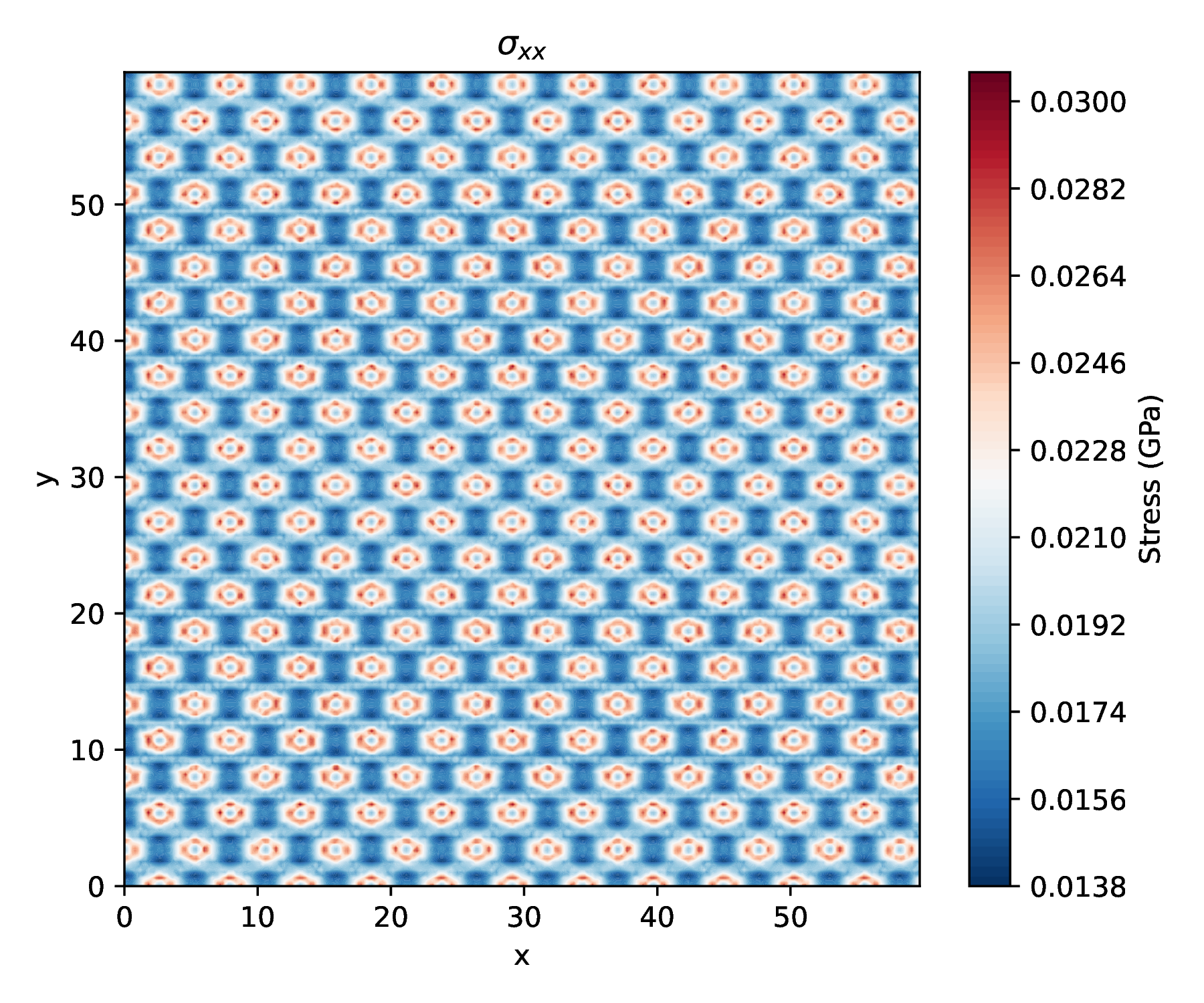
Virial Cauchy stress
[View high‑res PDF]
Full code: